FakET: Simulating Cryo-Electron Tomograms with Neural Style Transfer
Pavol Harar, Lukas Herrmann, Philipp Grohs, David Haselbach
Highlights:
• FakET adapted Neural style transfer technique to simulate cryo-electron micrographs
• It produces high-quality data resembling the unlabeled reference micrographs
• The simulation cost is far lower than that of complex physics-based simulators
• Offers a practical choice of simulating fully-labeled training data for deep learning
Description:
In cryo-electron microscopy, accurate particle localization and classification are imperative. Recent deep learning solutions, though successful, require extensive training data sets. The protracted generation time of physics-based models, often employed to produce these data sets, limits their broad applicability. Unfortunately, simulating the tomographic data using physics-based simulators is computationally very expensive. Since the protracted generation time limits their broad applicability, we introduce FakET [1], a method based on Neural Style Transfer [2], capable of expeditiously simulating the forward operator of any cryo transmission electron microscope. It can be used to adapt a synthetic training data set according to reference data producing high-quality simulated micrographs or tilt-series, see Figure 2. To assess the quality of our generated data, we used it to train DeepFinder [3], a state-of-the-art localization and classification architecture, and compared its performance with a counterpart trained on benchmark data generated by a physics-based simulator SHREC [4]. For more detailed comparison, see Table 1. Remarkably, our technique matches the performance, boosts data generation speed 750×, uses 33× less memory, and scales well to typical transmission electron microscope detector sizes. It leverages GPU acceleration and parallel processing. The source code is available at https://gitlab.com/deepet/faket.
[1] Harar, P., Herrmann, L., Grohs, P. and Haselbach, D., 2023. FakET: Simulating Cryo-Electron Tomograms with Neural Style Transfer. arXiv preprint arXiv:2304.02011.
[2] Gatys LA, Ecker AS, Bethge M. Image style transfer using convolutional neural networks. InProceedings of the IEEE conference on computer vision and pattern recognition 2016 (pp. 2414-2423).
[3] Moebel E, Martinez-Sanchez A, Lamm L, Righetto RD, Wietrzynski W, Albert S, Larivière D, Fourmentin E, Pfeffer S, Ortiz J, Baumeister W. Deep learning improves macromolecule identification in 3D cellular cryo-electron tomograms. Nature methods. 2021 Nov;18(11):1386-94.
[4] Gubins I, Chaillet ML, van Der Schot G, Veltkamp RC, Förster F, Hao Y, Wan X, Cui X, Zhang F, Moebel E, Wang X. SHREC 2020: Classification in cryo-electron tomograms. Computers & Graphics. 2020 Oct 1;91:279-89.
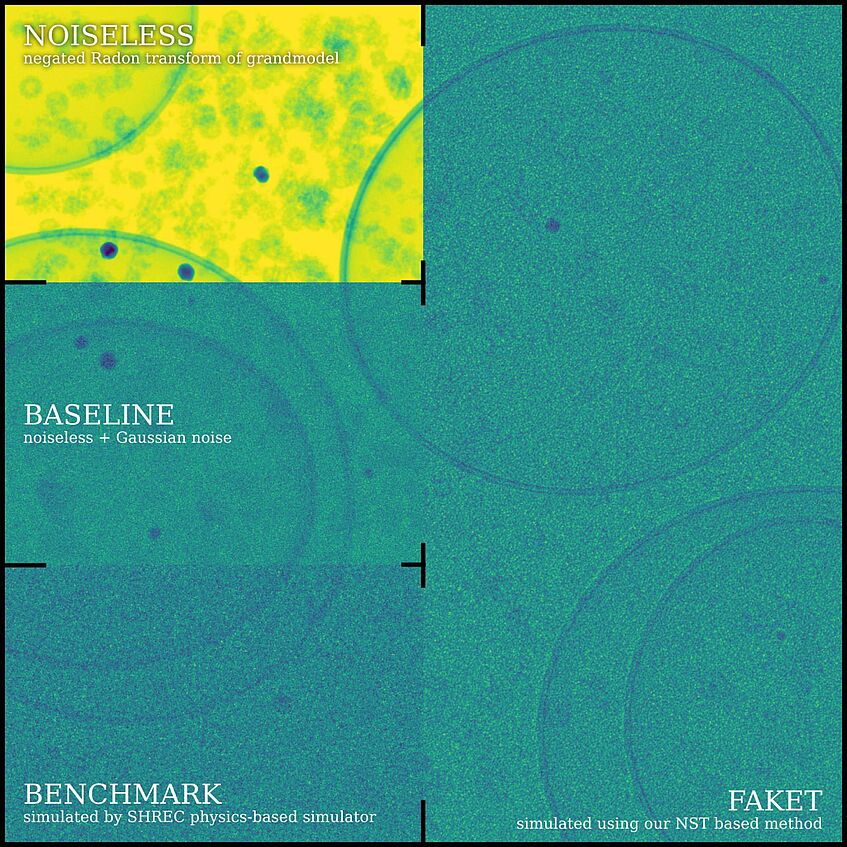
Figure 1: Side-by-side comparison of noiseless, baseline, benchmark, and FakET projections.
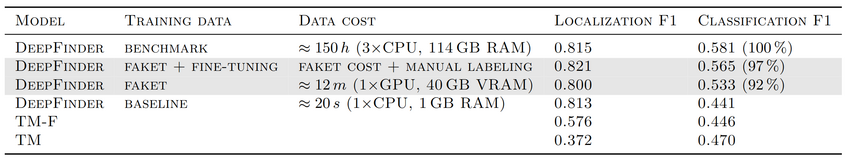
Table 1: Evaluation of simulated data quality by examining the performance on downstream tasks as a function of training data.
